Demo - RBIG¶
# @title Install Packages
# %%capture
try:
import sys, os
from pyprojroot import here
# spyder up to find the root
root = here(project_files=[".here"])
# append to path
sys.path.append(str(root))
except ModuleNotFoundError:
import os
os.system("pip install chex")
os.system("pip install git+https://github.com/IPL-UV/rbig_jax.git#egg=rbig_jax")
# jax packages
import jax
import jax.numpy as jnp
from jax.config import config
# import chex
config.update("jax_enable_x64", True)
import chex
import numpy as np
from functools import partial
# library functions
from rbig_jax.plots import plot_joint, plot_joint_prob, plot_info_loss
KEY = jax.random.PRNGKey(123)
# logging
import tqdm
import wandb
# plot methods
import matplotlib.pyplot as plt
from matplotlib import cm
import seaborn as sns
import corner
from IPython.display import HTML
%matplotlib inline
sns.reset_defaults()
sns.set_context(context="poster", font_scale=0.7)
%load_ext lab_black
%load_ext autoreload
%autoreload 2
INFO:tensorflow:Enabling eager execution
INFO:tensorflow:Enabling v2 tensorshape
INFO:tensorflow:Enabling resource variables
INFO:tensorflow:Enabling tensor equality
INFO:tensorflow:Enabling control flow v2
WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
Demo Data¶
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
# %%wandb
# get data
seed = 123
n_samples = 5_000
n_features = 2
noise = 0.05
X, y = datasets.make_moons(n_samples=n_samples, noise=noise, random_state=seed)
data = X[:]
# plot data
fig = corner.corner(data, color="blue", hist_bin_factor=2)
plt.show()
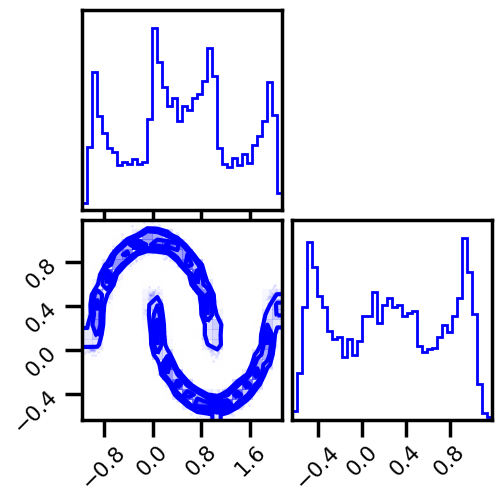
X = jnp.array(data, dtype=np.float64)
Model¶
from rbig_jax.models import RBIG
# a lot of parameters...
support_extension = 30
precision = 5_000
eps = 1e-5
method = "kde"
# Histogram specific params
nbins = None
alpha = 1e-5
# KDE specific params
bw = "scott"
# info reduction loss parameters
max_layers = 1_000
zero_tolerance = 60
p = 0.25
# initialize the max layers parameters
verbose = True
n_layers_remove = 50
interval = 10
# everything (makes it fast...)
jitted = True
Training¶
X_g, rbig_model = RBIG(
X=X,
support_extension=support_extension,
method=method,
precision=precision,
alpha=alpha,
nbins=nbins,
bw=bw,
eps=eps,
max_layers=max_layers,
zero_tolerance=zero_tolerance,
p=p,
verbose=verbose,
n_layers_remove=n_layers_remove,
interval=interval,
jitted=jitted,
)
Layer 10 - Cum. Info Reduction: 2.714 - Elapsed Time: 9.4249 secs
Layer 20 - Cum. Info Reduction: 3.182 - Elapsed Time: 16.3679 secs
Layer 30 - Cum. Info Reduction: 3.265 - Elapsed Time: 23.2687 secs
Layer 40 - Cum. Info Reduction: 3.413 - Elapsed Time: 30.2584 secs
Layer 50 - Cum. Info Reduction: 3.463 - Elapsed Time: 37.0513 secs
Layer 60 - Cum. Info Reduction: 3.463 - Elapsed Time: 44.5714 secs
Layer 70 - Cum. Info Reduction: 3.560 - Elapsed Time: 52.3466 secs
Layer 80 - Cum. Info Reduction: 3.611 - Elapsed Time: 59.3685 secs
Layer 90 - Cum. Info Reduction: 3.611 - Elapsed Time: 66.1504 secs
Layer 100 - Cum. Info Reduction: 3.611 - Elapsed Time: 72.9539 secs
Layer 110 - Cum. Info Reduction: 3.611 - Elapsed Time: 79.8892 secs
Layer 120 - Cum. Info Reduction: 3.700 - Elapsed Time: 86.6785 secs
Layer 130 - Cum. Info Reduction: 3.700 - Elapsed Time: 93.8044 secs
Layer 140 - Cum. Info Reduction: 3.769 - Elapsed Time: 100.7710 secs
Layer 150 - Cum. Info Reduction: 3.803 - Elapsed Time: 107.5936 secs
Layer 160 - Cum. Info Reduction: 3.803 - Elapsed Time: 114.4546 secs
Layer 170 - Cum. Info Reduction: 3.803 - Elapsed Time: 121.2034 secs
Layer 180 - Cum. Info Reduction: 3.875 - Elapsed Time: 128.1321 secs
Layer 190 - Cum. Info Reduction: 3.913 - Elapsed Time: 135.0527 secs
Layer 200 - Cum. Info Reduction: 3.913 - Elapsed Time: 142.0569 secs
Layer 210 - Cum. Info Reduction: 3.913 - Elapsed Time: 148.8200 secs
Layer 220 - Cum. Info Reduction: 3.913 - Elapsed Time: 155.7274 secs
Layer 230 - Cum. Info Reduction: 3.913 - Elapsed Time: 162.5874 secs
Layer 240 - Cum. Info Reduction: 3.913 - Elapsed Time: 169.3975 secs
Converged at Layer: 243
Final Number of layers: 193 (Blocks: 64)
Total Time: 171.3612 secs
Gaussianized Data¶
Training Loop¶
# plot data
fig = corner.corner(np.array(X_g), color="red", hist_bin_factor=2)
plt.show()
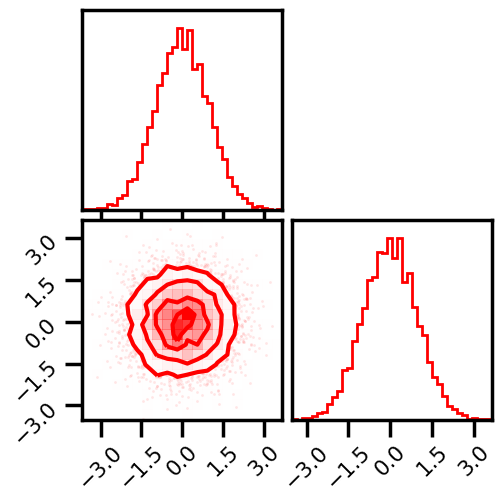
From Model¶
%%time
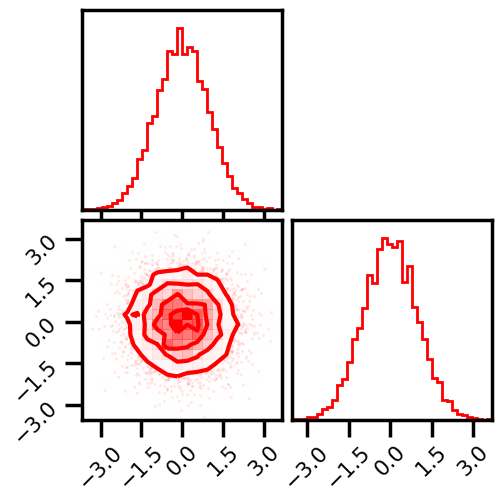
X_g_ = rbig_model.forward(X)
# plot data
fig = corner.corner(np.array(X_g_), color="red", hist_bin_factor=2)
plt.show()
CPU times: user 18.6 s, sys: 1.34 s, total: 20 s
Wall time: 17.1 s
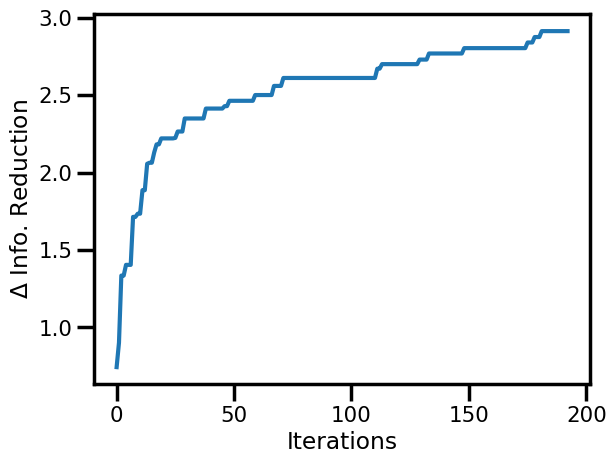
Information Reduction Evolution¶
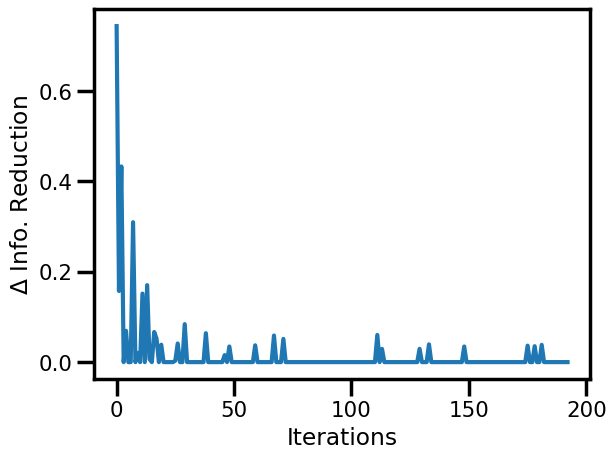
fig, ax = plt.subplots()
ax.plot(rbig_model.info_loss)
ax.set(xlabel="Iterations", ylabel="$\Delta$ Info. Reduction")
plt.show()
fig, ax = plt.subplots()
ax.plot(jnp.cumsum(rbig_model.info_loss))
ax.set(xlabel="Iterations", ylabel="$\Delta$ Info. Reduction")
plt.show()
Negative Log-Likelihood¶
%%time
X_valid, y = datasets.make_moons(n_samples=1_000, noise=noise, random_state=42)
nll = rbig_model.score(X_valid)
print(f"NLL Score: {nll:.4f}")
NLL Score: 0.9947
CPU times: user 35.4 s, sys: 2.46 s, total: 37.9 s
Wall time: 34.4 s
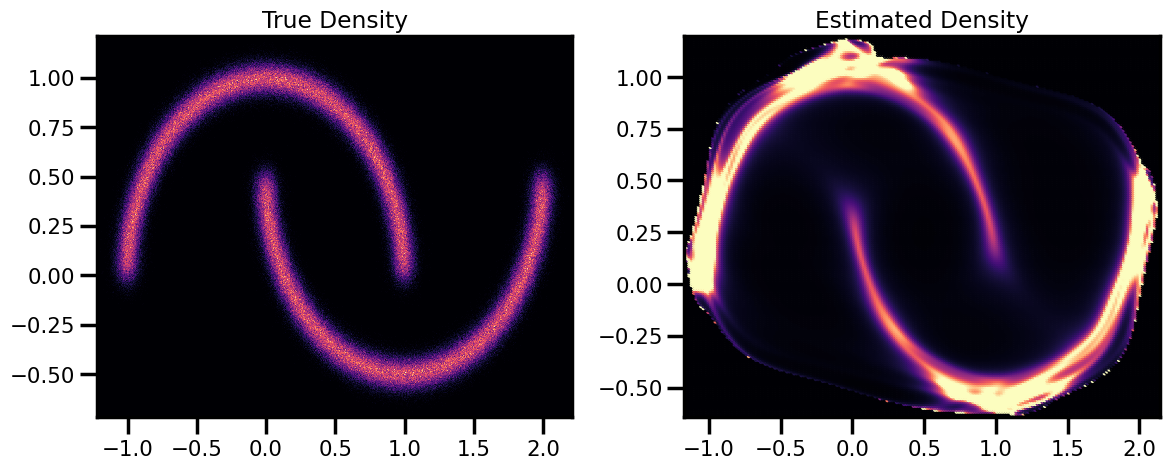
Density Estimation¶
# Original Density
n_samples = 1_000_000
noise = 0.05
seed = 42
X_plot, _ = datasets.make_moons(n_samples=n_samples, noise=noise, random_state=seed)
%%time
n_grid = 200
buffer = 0.01
xline = jnp.linspace(X[:, 0].min() - buffer, X[:, 0].max() + buffer, n_grid)
yline = jnp.linspace(X[:, 1].min() - buffer, X[:, 1].max() + buffer, n_grid)
xgrid, ygrid = jnp.meshgrid(xline, yline)
xyinput = jnp.concatenate([xgrid.reshape(-1, 1), ygrid.reshape(-1, 1)], axis=1)
X_log_prob = rbig_model.score_samples(xyinput)
CPU times: user 57.5 s, sys: 3.31 s, total: 1min
Wall time: 42.9 s
Generate Grid Points¶
Plot - Compare¶
# Estimated Density
cmap = cm.magma # "Reds"
probs = jnp.exp(X_log_prob)
fig, ax = plt.subplots(ncols=2, figsize=(12, 5))
h = ax[0].hist2d(
X_plot[:, 0], X_plot[:, 1], bins=512, cmap=cmap, density=True, # vmin=0.0, vmax=1.0
)
ax[0].set_title("True Density")
ax[0].set(
xlim=[X_plot[:, 0].min(), X_plot[:, 0].max()],
ylim=[X_plot[:, 1].min(), X_plot[:, 1].max()],
)
h1 = ax[1].scatter(
xyinput[:, 0], xyinput[:, 1], s=1, c=probs, cmap=cmap, vmin=0.0, vmax=1.0
)
ax[1].set(
xlim=[xyinput[:, 0].min(), xyinput[:, 0].max()],
ylim=[xyinput[:, 1].min(), xyinput[:, 1].max()],
)
# plt.colorbar(h1)
ax[1].set_title("Estimated Density")
plt.tight_layout()
plt.show()
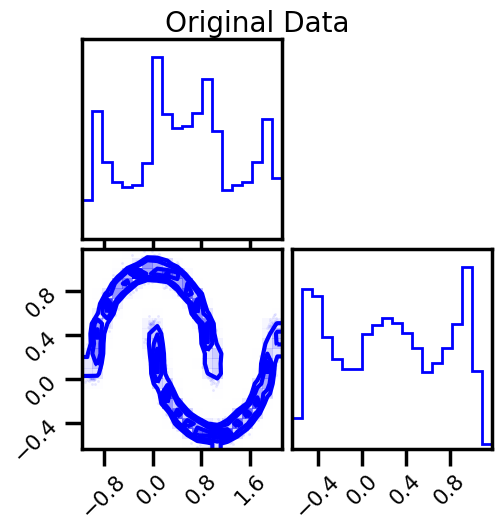
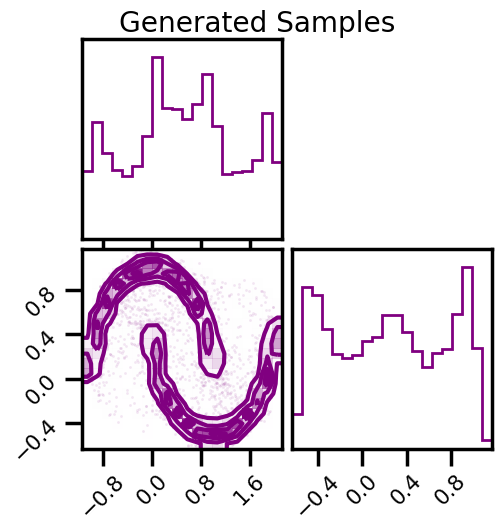
Sampling¶
%%time
# number of samples
n_samples = 10_000
seed = 42
X_samples = rbig_model.sample(seed=seed, n_samples=n_samples)
CPU times: user 18.5 s, sys: 1.12 s, total: 19.7 s
Wall time: 16.3 s
fig = corner.corner(np.array(X), color="blue", label="Original Data")
fig.suptitle("Original Data")
plt.show()
fig2 = corner.corner(np.array(X_samples), color="purple")
fig2.suptitle("Generated Samples")
plt.show()
Saving and Loading¶
Often times it would be nice to save and load models. This is useful for checkpointing (during training) and also for convenience if you’re doing research on google colab.
Fortunately, everything here are python objects, so we can easily save and load our models via pickle.
Saving¶
Do to the internals of python (and design choices within this library), one can only store objects. So that includes the rbig_block, the bijectors and also the rbig_model. This does not include the rbig_block_init for example because that isn’t an object, it’s a function with some local params.
import joblib
joblib.dump(rbig_model, "rbig_model_plane.pickle")
['rbig_model_plane.pickle']
Simple Test¶
They won’t be the exact same byte-for-byte encoding. But they should give the same results either way :).
# nll for the old model
nll = rbig_model.score(X)
# nll for the loaded model
nll_loaded = rbig_model_loaded.score(X)
# check that they're the same
chex.assert_tree_all_close(nll, nll_loaded)
print(f"NLL (Original): {nll:.4f}")
print(f"NLL (Loaded): {nll_loaded:.4f}")
NLL (Original): 0.5863
NLL (Loaded): 0.5863